MIT designs circuits based on protein-protein interactions
However, there is often a long lag time between an event such as detecting a molecule and the resulting output, because of the time required for cells to transcribe and translate the necessary genes.
MIT synthetic biologists have now developed an alternative approach to designing such circuits, which relies exclusively on fast, reversible protein-protein interactions. This means that there’s no waiting for genes to be transcribed or translated into proteins, so circuits can be turned on much faster — within seconds.
“We now have a methodology for designing protein interactions that occur at a very fast timescale, which no one has been able to develop systematically. We’re getting to the point of being able to engineer any function at timescales of a few seconds or less,” says Deepak Mishra, a research associate in MIT’s Department of Biological Engineering and the lead author of the new study.
This kind of circuit could be useful for creating environmental sensors or diagnostics that could reveal disease states or imminent events such as a heart attack, the researchers say.
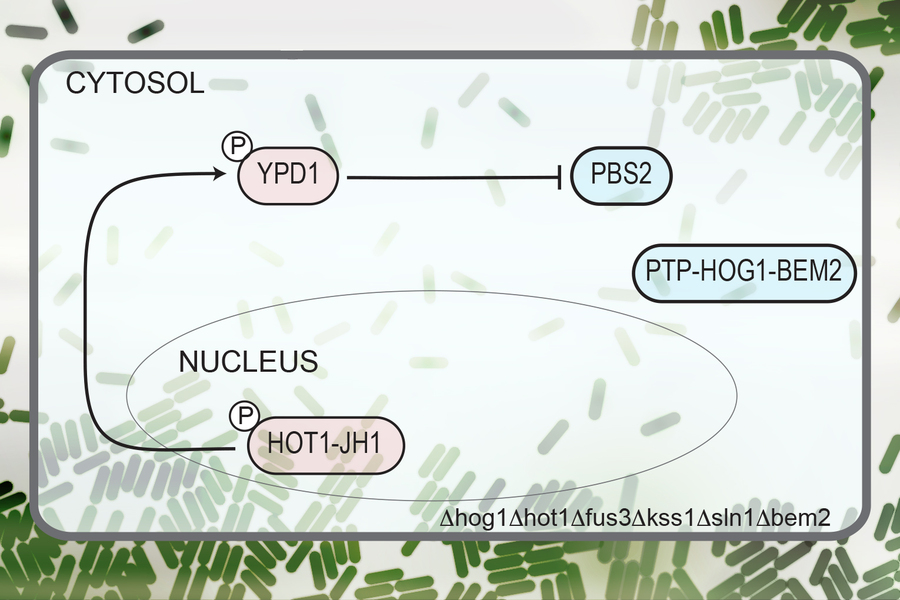
Inside living cells, protein-protein interactions are essential steps in many signaling pathways, including those involved in immune cell activation and responses to hormones or other signals. Many of these interactions involve one protein activating or deactivating another by adding or removing chemical groups called phosphates.
In this study, the researchers used yeast cells to host their circuit and created a network of 14 proteins from species including yeast, bacteria, plants, and humans. The researchers modified these proteins so they could regulate each other in the network to yield a signal in response to a particular event.
Their network, the first synthetic circuit to consist solely of phosphorylation / dephosphorylation protein-protein interactions, is designed as a toggle switch — a circuit that can quickly and reversibly switch between two stable states, allowing it to “remember” a specific event such as exposure to a certain chemical. In this case, the target is sorbitol, a sugar alcohol found in many fruits.
Once sorbitol is detected, the cell stores a memory of the exposure, in the form of a fluorescent protein localized in the nucleus. This memory is also passed on to future cell generations. The circuit can also be reset by exposing it to a different molecule, in this case, a chemical called isopentenyl adenine.
These networks can also be programmed to perform other functions in response to an input. To demonstrate this, the researchers also designed a circuit that shuts down cells’ ability to divide after sorbitol is detected.
By using large arrays of these cells, the researchers can create ultrasensitive sensors that respond to concentrations of the target molecule as low as parts per billion. And because of the fast protein-protein interactions, the signal can be triggered in as little as one second. With traditional synthetic circuits, it could take hours or even days to see the output.